
Associate Professor
Biochemistry
Ph.D., Oregon Health Sciences University, 2000
Tel: 316.978.7373 (office)
316.978.7322 (lab)
Fax: 316.978.3431
Email: Jim.Bann@wichita.edu
Research:
Anthrax Protective Antigen
The anthrax protective antigen (PA) is an 83 kDa protein secreted by Bacillus anthracis, the causative agent of anthrax disease. Upon infection by B anthracis, PA and two other proteins, edema factor (EF) and lethal factor (LF) are secreted from the bacterium into the host bloodstream. PA binds to host cellular receptors, and after proteolytic processing assembles into heptameric (Lacy et al., PNAS 2004) or octameric (Kintzer et al., JMB 2009) ring-shaped structures called pre-pores. These pre-pore structures allow EF and LF to bind, and the entire complex is endocytosed into the host cell. Eventually, the resulting endosome becomes acidified as it is trafficked along, and forms a membrane spanning pore. The structure of the pore has recently been visualized in stunning detail using cryo-EM (Jiang et al., Nature 2015). Our research is directed towards understanding the mechanistic details of how PA forms a membrane spanning pore at low pH. Formation of the pore is absolutely critical for the pathogenesis of the toxin, since the pore provides a conduit for entry of the edema or lethal factor into the cell cytosol.
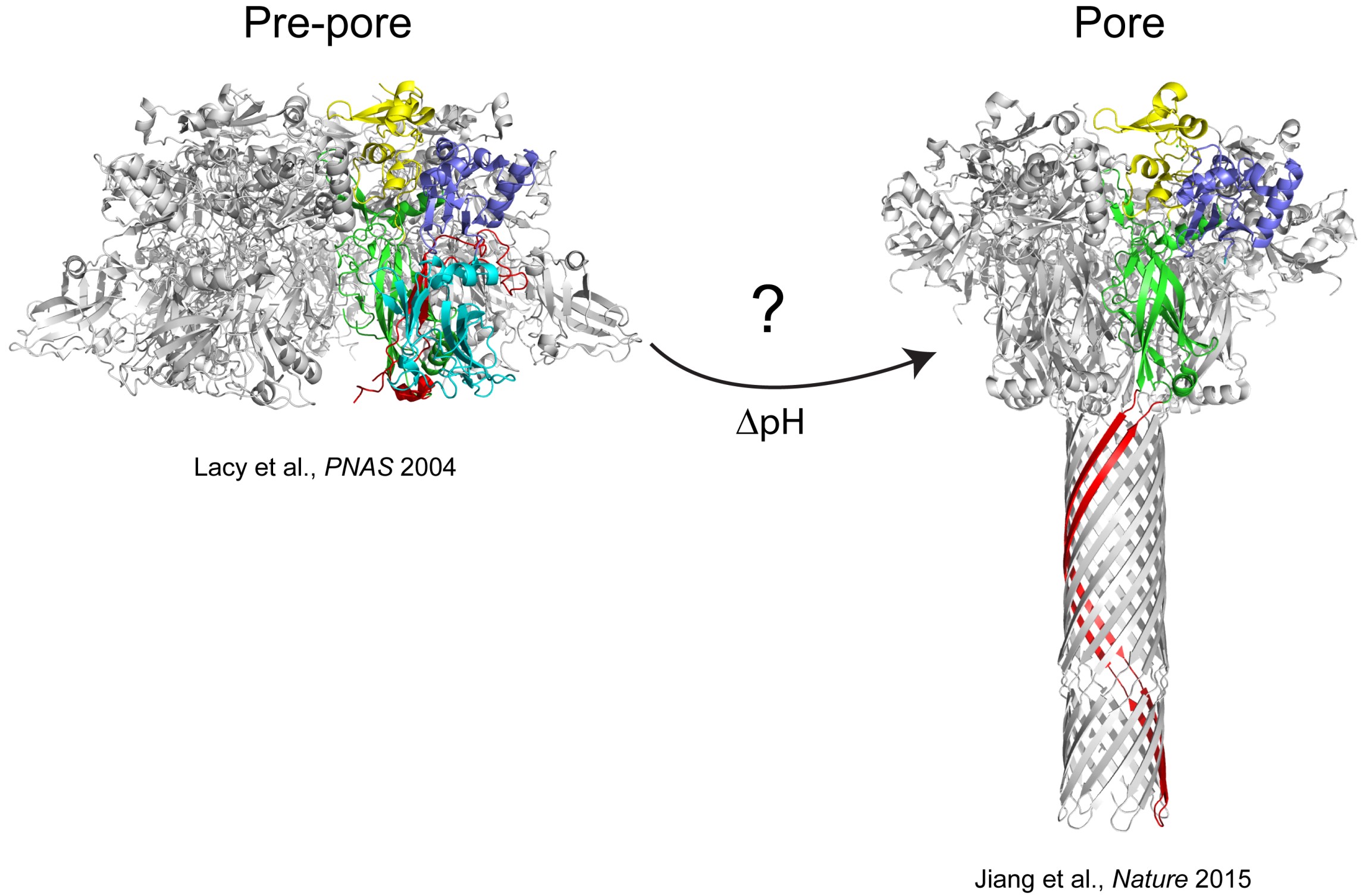
Although the exact mechanism of pore formation is not understood, seminal work from
the Collier laboratory at Harvard has shown that pore formation can be prevented by
mutating certain amino acid residues within PA (dominant-negative mutants, Sellman
et al., JBC 2001). We are currently investigating how these and other mutations prevent pore
formation, which may provide clues as to the pathway(s) needed for this large structural
change.
The methods we use include circular dichroism spectroscopy, fluorescence (equilibrium
and stopped-flow) and fluorine NMR. We have also made use of mutagenesis as well as
incorporation of unnatural amino acids to investigate the mechanism of pore formation.
In the latter case for instance, we biosynthetically incorporated into PA 2-fluorohistidine,
which has a significantly lower pKa than histidine (pKa of ~1). Biosynthetic incorporation
does not prevent pore formation of PA unless the soluble form of the host cellular
receptor capillary morphogenesis protein 2 (sCMG2) is bound(1). This suggested that
histidine protonation could be a trigger for pore formation in the context of the
receptor bound state, and lead us to investigate further differences in structure
between the receptor bound and unbound states(2).
In collaboration with Masaru Miyagi (Case Western Reserve), we have shown that the thermodynamic stability of PA is greatly increased if sCMG2 is bound (2, 3), and occurs throughout the protein structure. Give the increased stability of PA, and the fact that it is a well-studied immunogen, we are also pursuing studies directed at understanding the role of protein stability in immunogenicity.
Selected Publications:
1. Wimalasena, D.S., Janowiak, B.E., Lovell, S., Miyagi, M., Sun, J., Zhou, H., Hajduch,
J., Pooput, C., Kirk, K.L., Battaile, K.P., and Bann, J.G. (2010). Evidence that histidine
protonation of receptor-bound anthrax protective antigen is a trigger for pore formation.
Biochemistry 49, 6973-6983.
2. Mullangi, V., Mamillapalli, S., Anderson, D.J., Bann, J.G., and Miyagi, M. (2014).
Long-range stabilization of anthrax protective antigen upon binding to CMG2. Biochemistry, 38, 6084-6091.3.
3. Chadegani, F., Lovell, S., Mullangi, V., Miyagi, M., Battaile, K.P., and Bann,
J.G. (2014). 19F nuclear magnetic resonance and crystallographic studies of 5-fluorotryptophan-labeled
anthrax protective antigen and effects of receptor on stability. Biochemistry, 53, 690-701.
Fluorine NMR

Our laboratory has developed methods to biosynthetically incorporate fluorinated amino acids into proteins, and a major focus of our laboratory is to expand the repertoire of fluorinated amino acids that have unique functionalities, and to incorporate these amino acids into proteins for structural studies. Fluorine (19F), is only slightly larger than hydrogen, is a spin ½ nucleus, is 100% naturally abundant and is almost as sensitive as 1H. The NMR spectrum of a fluorinated protein typically exhibits well resolved resonances which are sensitive to environment, allowing the researcher to follow structurally changes in select regions of the protein at a residue-specific level. However, the origin of the fluorine chemical shift in the context of proteins is not well understood. In collaboration with the Mitchell-Koch laboratory, we are investigating the origin of fluorine chemical shifts beginning with simple amino acids and progressing from there to peptides and proteins.
Selected Publications:
Eichler, J.F., Cramer, J.C., Kirk, K.L., and Bann, J.G. (2005). Biosynthetic Incorporation
of Fluorohistidine into Proteins in E.coli: A New Probe of Macromolecular Structure.
ChemBioChem. 6, 1-4.
Thomas, C.A., Talaty, E.R., and Bann, J.G. (2009). 3S-fluoroproline as a probe to
monitor proline isomerization during protein folding by 19F-NMR. Chem. Comm. 23, 3366-3368.
Andra, K.K., Bullinger, J.C., Bann, J.G., and Eichhorn, D.M. (2010). 2-Fluoro-L-histidine.
Acta Crystallogr Sect E Struct Rep Online. 66, 2713.
Kasireddy, C., Bann, J.G., and Mitchell-Koch, K.R. (2015). Demystifying fluorine
chemical shifts: electronic structure calculations address origins of seemingly anomalous
19F-NMR spectra of fluorohistidine isomers and analogues. Phys Chem Chem Phys. 45, 30606-30612.
Kasireddy, C., Ellis, J.M., Bann, J.G., Mitchell-Koch, K.R. (2017). The Biophysical
Probes 2-fluorohistidine and 4-fluorohistidine:Spectroscopic Signatures and Molecular
Properties. Sci. Rep. 7, srep42651(online).

